大数据集病理切片图像的分析(分类,分割,聚类)
Parallel Multiple Instance Learning forExtremely LargeHistopathology ImagesAnalysis
(本项目受微软亚洲研究院资助)
(This project was funded by Microsoft Research Asia)
INTRODUCTION
Cancer disease is one of the principle causes which lead to death in the world. The detection methods for cancer disease are some such as PET, CT, MRI and histopathology. Only histopathology of a rich source of information in high resolution can completely and correctly detect cancer disease. For recent years, computers dramatically increase in their computational power, speed and storage capability digitalization of tissue histopathology is now possible through scanning tissue sections using a specialized digital microscope scanner. The data size obtained on a digitalized whole slide tissue histopathological image is enormous. For example, a digitalized histopathological image at 40 x resolution is the size of roughly 15,000 x 15,000 pixels. It is common that pathology section processing generates 12-20 images for each patient. It is only now that the digitalized histopathology datasets dramatically increase. With the development of computers, histopathological image analysis from cancer tissue becomes a hot topic. Firstly, automated quantitative analysis with computer assisted diagnosis (CAD) for cancer tissue based on whole slide histopathological images will improve the accuracy for cancer grading. The cancer grading is depending on the gland shape and size. It is difficult that the gland shape and size are computed through the pathologists’ observation. It is easier for computers to complete the exact quantification for the gland shape and size. Secondly, the analysis will save a considerable amount of resources a manual labour. As far as prostate cancer, there are approximately 1 million biopsies performed in US each year. Only 20% are found to belong to cancer. Even if, the automated quantity analysis can detect malignant or benign, pathologists can save 80% time to observe the malignant parts in more details. In addition, for the beginners of the pathologists, they need to accumulate a huge mass of experience through observing the resultful histopathological images. The system can train and teach them what they want. There are a lot of cancers in various organs including lung, prostate, breast and colon. Without loss of generality, we choose colon cancer as a case study. We propose to annotate some colon histopathological data as research use and collaborate with Microsoft Research Asia to develop a platform of automated quantitative analysis with CAD in cancer histopathological images and apply it to clinical practice.
ACHIEVEMENT
First, the system can automatically detect cancer, segment cancer regions and cluster cancer type (See Figure 1).
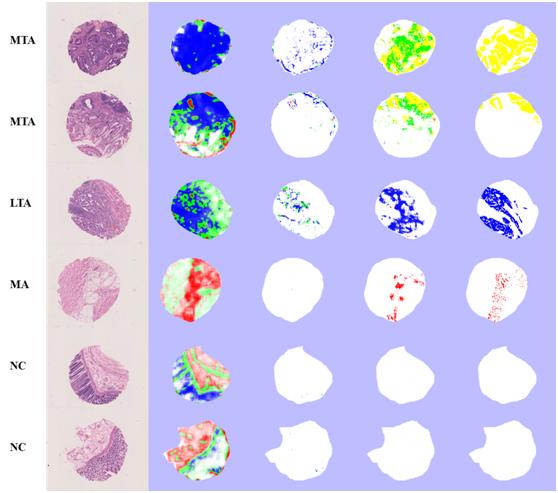
Figure 1: Image Types: (a): The original images. (b), (c), (d): The instance-level results (pixel-level segmentation and patch-level clustering) for standard Boosting + K-means, MIL + K-means, and our MCIL. (e): The instance-level ground truth labeled by three pathologists. Different colors stand for different types of cancer tissues. Cancer Types: from top to bottom: MTA, MTA, LTA, MA, NC, and NC. [Five types of colon cancer images are used: Non-cancer (NC), Middle tubular adenocarcinoma (MTA), Low tubular adenocarcinoma (LTA), Mucinous adenocarcinoma (MA), and Signet-ring carcinoma (SRC)].
Second, in order to train on large data sets ina reasonable amount of time, we developed parallel multiple instance learning(P-MIL) algorithm on High-Performance-Computing (HPC) clusters, using acombination of Message Passing Interface (MPI) and multi-threading.Ourexperiments are conducted on a Microsoft Windows HPC cluster, which is ahomogeneous infrastructure consisting of 128 compute nodes, connected by network with high bandwidth and low latency.Each compute node has 2 quad-coreXeon 2.43-GHz processors, 16GB RAM, 1Gbit Ethernet adapters and 1.7TBlocal disk storage.Figure 2 is the flowdiagram for the parallel multiple instancelearning.We then successfully conducted a thorough experiment study inwhich our model is trained using millions instances and 215 features each in 128compute nodes (1024 CPU cores) for 25.1 hours.The dataset is 6.135 trillionbyte colon cancer images in our experiment.

PUBLICATION
Yan Xu, Jun-Yan Zhu, Eric Chang, and Zhuowen Tu, Multiple Clustered Instance Learning for Histopathology Cancer Image Classification, Segmentation and Clustering, in Computer Vision and Pattern Recognition (CVPR), 2012
Yan Xu, Jianwen Zhang, Eric Chang, Maode Lai, and Zhuowen Tu, Contexts-Constrained Multiple Instance Learning for Histopathology Image Analysis (Oral), in International Conference on Medical Image Computing and Computer Assisted Intervention (MICCAI), June 2012